|
|
Application Example
CAT1/CAT2 identification in Solanum nigrum L.
The usability of PrimerIdent was tested by designing primers
that could be used to identify different different isozymes
of the antioxidative plant defence system enzyme catalase (CAT)
from Solanum nigrum L.
CAT is one of the main H2O2 catalysts and has several isozymes
grouped according to their cellular localization: CAT1 – mainly
expressed in photosynthetic tissues; CAT2 – essentially expressed
in vascular tissues; CAT3 – expressed in seeds and young plants
(Willekens et al., 1994).
S. nigrum, commonly known as black nightshade, is a plant species
with very few described nucleotide sequences. Due to its phylogenetic
proximity to Potato (Solanum tuberosum), tomato (Solanum
lycopersicum), chilli-pepper (Capsicum annum) and
tobacco (Nicotiana tabaccum ), cDNAs from these plant
species were used to assemble the sequences alignments for each
type of enzyme to be studied:
[››]S. tuberosum CAT1 (AY442179),
S. lycopersicum CAT1 (AF112368), C. annum CAT1
(AY128694), N. tabacum CAT1 (NTU93244),
[››]S. tuberosum CAT2 (AY500290),
S. lycopersicum CAT2 (M93719), C. annum CAT2 (AB007190)
and N. tabacum CAT2 (NTU07627).
CAT3 wasn’t included in the analysis due to the lack of sequence
information that would ensure a reliable and specific primer
design.
The FASTA alignments were assembled for each case and submitted
to PrimerIdent for left primer search, using default settings,
with the exception of GC-Clamp, which was changed to 1 (all
primers found end in either a G or a C). The sequence chosen
for template was from S. tuberosum. The different primers chosen
for each of the cDNAs studied are depicted in Table 1.
|
Primer name |
Primer sequence |
Identity for each
of the alignment sequences (%) |
|
Solanum tuberosum |
Solanum lycopersicum |
Capsicum annum |
Nicotiana tabaccum |
|
CAT1 |
CTGCCCTTCTATTGTGGTTC |
100
(CAT1) |
45,0
(CAT2) |
100
(CAT1) |
45,0
(CAT2) |
100
(CAT1) |
45,0
(CAT2) |
100
(CAT1) |
45,0
(CAT2) |
|
CAT2 |
CGTTCCTTCCTGAGAGTTTG |
100
(CAT2) |
55,0
(CAT1) |
100
(CAT2) |
55,0
(CAT1) |
85,0
(CAT2) |
60,0
(CAT1) |
90,0
(CAT2) |
60,0
(CAT1) |
Table 1 - Left primers chosen from the PrimerIdent reports
3' RACE
The forward (left) primers (Table 1) chosen with the help of
PrimerIdent were successfully used in 3’ RACE reactions,
along with a reverse primer-adapter for CAT1 and CAT2 (Figure
1). Each RACE reaction for each primer produced one band
that was cloned and sequenced. The sequences obtained corresponded
to each of the enzymes expected and, therefore, were published
in NCBI. As a result, PrimerIdent helped selecting specific
PCR primers that led to the successful identification of
the following partial sequences:
[››]FJ402837 – Solanum nigrum
CAT1;
[››]FJ402838 – Solanum nigrum
CAT2;
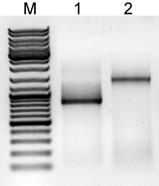
Figure 1 - Agarose gel electrophoresis for the 3’ RACE reactions
M – GeneRuller DNA ladder Mix (Fermentas, Lithuania), 1
µg;
1 – 3’ RACE for S. nigrum CAT1;
2 – 3’ RACE for S. nigrum CAT2.
Report Analysis
Table 2 depicts the statistical analysis from a PrimerIdent report
and shows how conserved primers across multiple species and
simultaneously specific for a certain isozyme can be rare. Of
all 409 primers found (using the default conditions of PrimerIdent
and using S. tuberosum CAT1 as template sequence) only 73 (23%)
are conserved across all 4 Solanaceae. A more relaxed criterion
than strict conservation can be used to define a primer suitable
for S. nigrum: 100% identity for Potato/Tomato and >85%
identity for Tobacco and Pepper. Choosing primers with this
criterion increases the possible primers to 241 (59%). However,
when taking into account that these primers must have a low
identity to CAT2 sequences in order to be specific to CAT1,
the number of primers found drops to only 34 (8% of the total
primers). It should be emphasized that the primers provided
in the final report are not degenerated. The researcher can
choose from the resulting list the primer that presents the
maximum identity for the target cDNA, while presenting the lowest
identity for related nucleotide sequences.
|
|
Total primers |
Conserved primers |
Primers suitable for
S. nigrum |
Primers suitable for
S. nigrum with low identity to CAT2 |
|
S. tuberosum
CAT1 as template
sequence |
409 |
73 |
241 |
34 |
Table 2 - Statistical results from a PrimerIdent analysis
Figure 2 shows the analysis of the distribution of the conserved
primers across the Potato CAT1 sequence showing that the 28
CAT1-specific primers with an identity lower than 60% to the
correspondent sequences of CAT2 are clustered in only 8 different
regions of the sequence which further demonstrates how difficult
would be to find them using the trial-error method. 7770

Figure 2 - Location of the primers described in Table 2 across
S. tuberosum CAT1 sequence (AY442179) The numbers represent
the sequence length (in bp);
● represents the location of the primers considered
suitable for CAT1 of S. nigrum;
▲ represents the location of the conserved primers;
♦ represents the location
of the primers considered suitable for CAT1 of S. nigrum
with an identity lower than 60% to the correspondent sequences
of CAT2.
|
| Copyright Notice |
|
PrimerIdent is a Copyright (c) of Alberto Pessoa,
Faculdade de Ciências - Universidade
do Porto, Portugal; 2009-2013.
PrimerIdent uses Primer3, Copyright (c) 1996,1997,1998,1999,2000,2001,2004,2006,2007,2008,2009,2010,2011,2012
Whitehead Institute for Biomedical Research,
Steve Rozen, and Helen Skaletsky
All rights reserved.
|
|


